-Search query
-Search result
Showing 1 - 50 of 131 items for (author: baek & k)

EMDB-18609: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0
Method: single particle / : Kovalev K, Podoliak E, Lamm GHU, Marin E, Stetsenko A, Guskov A

EMDB-18610: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3
Method: single particle / : Kovalev K, Podoliak E, Lamm GHU, Marin E, Stetsenko A, Guskov A

PDB-8qqz: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 8.0
Method: single particle / : Kovalev K, Podoliak E, Lamm GHU, Marin E, Stetsenko A, Guskov A

PDB-8qr0: 
Cryo-EM structure of the light-driven sodium pump ErNaR in the pentameric form at pH 4.3
Method: single particle / : Kovalev K, Podoliak E, Lamm GHU, Marin E, Stetsenko A, Guskov A

EMDB-43226: 
DNA origami colloid for self-assembly of tubules: (6,0) monomer
Method: single particle / : Videbaek TE, Rogers WB

EMDB-43227: 
DNA origami colloid for self-assembly of tubules: (10,0) monomer
Method: single particle / : Videbaek TE, Rogers WB

EMDB-16891: 
Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6
Method: single particle / : Wallace I, Kheewoong B, Prabu JR, Vollrath R, von Gronau S, Schulman BA, Swatek KN
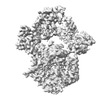
EMDB-18589: 
Map of UBE1L activating enzyme bound to ISG15 (FL) and UBE2L6
Method: single particle / : Wallace I, Kheewoong B, Prabu JR, Vollrath R, von Gronau S, Schulman BA, Swatek KN

PDB-8oif: 
Structure of the UBE1L activating enzyme bound to ISG15 and UBE2L6
Method: single particle / : Wallace I, Kheewoong B, Prabu JR, Vollrath R, von Gronau S, Schulman BA, Swatek KN
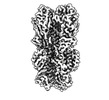
EMDB-40557: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
Method: single particle / : Borst AJ, Bennett NR

PDB-8sk7: 
Cryo-EM structure of designed Influenza HA binder, HA_20, bound to Influenza HA (Strain: Iowa43)
Method: single particle / : Borst AJ, Bennett NR

EMDB-40162: 
E. coli MlaC bound to MlaFEDB
Method: single particle / : MacRae MR, Coudray N, Ekiert D, Bhabha G

EMDB-28858: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

EMDB-28859: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

EMDB-28860: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

PDB-8f4x: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

PDB-8f53: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

PDB-8f54: 
Top-down design of protein architectures with reinforcement learning
Method: single particle / : Borst AJ, Baker D

EMDB-14561: 
CAND1-CUL1-RBX1
Method: single particle / : Baek K, Schulman BA

EMDB-14563: 
CAND1-SCF-SKP2 CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14564: 
CAND1-SCF-SKP2 (SKP1deldel) CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14594: 
CAND1-SCF-SKP2 CAND1 rolling-2 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14597: 
CAND1 delhairpin-SCF-SKP2 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14598: 
CAND1 delhairpin-SCF-SKP2 CAND1 rolling SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14599: 
CAND1-SCF-FBXW7 CAND1 rolling-1a
Method: single particle / : Baek K, Schulman BA

EMDB-14600: 
CAND1-SCF-FBXW7 CAND1 rolling-1b
Method: single particle / : Baek K, Schulman BA

EMDB-14601: 
CAND1-SCF-FBXW7 CAND1 rolling-2c
Method: single particle / : Baek K, Schulman BA

EMDB-14602: 
CAND1-SCF-FBXW7 CAND1 rolling-2d
Method: single particle / : Baek K, Schulman BA

EMDB-14603: 
CAND1-SCF-FBXW7 CAND1 rolling-2a
Method: single particle / : Baek K, Schulman BA

EMDB-14604: 
CAND1-SCF-FBXW7 CAND1 rolling-2b
Method: single particle / : Baek K, Schulman BA

EMDB-14605: 
CAND1-SCF-FBXW7 CAND1 little CAND1
Method: single particle / : Baek K, Schulman BA

EMDB-14606: 
CAND1-SCF-FBXW7 CAND1 no CAND1-1
Method: single particle / : Baek K, Schulman BA

EMDB-14607: 
CAND1-SCF-FBXW7 CAND1 no CAND1-2
Method: single particle / : Baek K, Schulman BA

EMDB-14608: 
CAND1-SCF-FBXW7 (SKP1deldel) CAND1 engaged no SKP1-Fbp
Method: single particle / : Baek K, Schulman BA

EMDB-14609: 
CAND1-SCF-FBXW7 (SKP1deldel) CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14610: 
CAND1delhairpin-SCF-FBXW7 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA

EMDB-14611: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2d SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14612: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2c SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14613: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2b SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14614: 
CAND1delhairpin-SCF-FBXW7 CAND1 rolling-2a SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14615: 
CAND1(2X)-SCF-FBXW7 no CAND1 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-14616: 
CAND1(2X)-SCF-FBXW7 CAND1 rolling-1 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-16575: 
CAND1 b-hairpin++-SCF-SKP2 CAND1 rolling SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-16576: 
CAND1 b-hairpin++-SCF-SKP2 CAND1 partly engaged SCF partly rocked
Method: single particle / : Baek K, Schulman BA

EMDB-16577: 
CAND1(2X)-SCF-FBXW7 CAND1 rolling-2 SCF engaged
Method: single particle / : Baek K, Schulman BA

EMDB-16578: 
CAND1(2X)-SCF-FBXW7 CAND1 engaged SCF rocked
Method: single particle / : Baek K, Schulman BA

EMDB-16579: 
CAND1-SCF-FBXW7 no CAND1 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA

EMDB-16580: 
CAND1-SCF-FBXW7 CAND1 rolling-1 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA

EMDB-16581: 
CAND1-SCF-FBXW7 CAND1 rolling-2 SCF engaged (low res)
Method: single particle / : Baek K, Schulman BA

EMDB-16582: 
CAND1-SCF-SKP2 CAND1 rolling-1 SCF engaged
Method: single particle / : Baek K, Schulman BA
Pages:
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
